Abstract
Type 2 diabetes mellitus (T2DM), a hyperglycemic metabolic condition, is considered one of the leading causes of mortality worldwide. Sirtuin 1 (SIRT1), a protein encoder gene, is involved in the protection of pancreatic β-cells from oxidative stress as part of its homeostatic functions. Mutation of the SIRT1 gene has been reported to be associated with T2DM in different populations. Therefore, this study aimed to reveal the association of the rs3758391 polymorphism with T2DM occurrence in the Appalachian population. For this study, 38 T2DM patients and 43 healthy controls were recruited, and genotyping of the SNP was performed by polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP). The CC genotype yielded a band of 241 bp, the TT genotype yielded bands of 148 bp and 93 bp, while the CT genotype yielded bands of 241 bp, 148 bp, and 93 bp. Data from our small-scale genome-wide association study (GWAS) between rs3758391 polymorphism and increased risk of T2DM did not show any significant relationship under co-dominant TT vs CC (OR=2.625, 95% CI=0.46-15.11, P=0.27) & CT vs CC (OR=1.26, 95% CI=0.50-3.15, P=0.62), recessive TT vs CC+CT (OR= 2.368, 95% CI= 0.43-12.99, P= 0.31) and allelic T vs C (OR=1.44, 95% CI=0.72-2.88, P= 0.29) genetic models.
Graphical Abstract
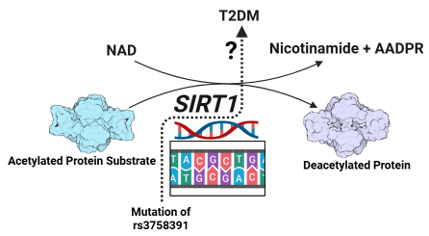
Keywords: Type 2 diabetes mellitus (T2DM), SIRT1 gene
Introduction
Diabetes mellitus (DM), a metabolic disorder characterized by hyperglycemia, is one of the leading causes of illness and mortality worldwide as a result of microvascular and macrovascular complications (1,2). Type 2 diabetes mellitus (T2DM) constitutes the majority of diabetes cases, representing 96% of the overall diabetes prevalence in 2021, and is heavily influenced by genetic factors and closely linked to obesity and a physically inactive lifestyle (2,3). The Appalachian area is largely rural and culturally distinct, with unique economic and healthcare challenges from the rest of the nation. In addition to these characteristics, an inadequate number of health professionals in this region might contribute to the prevalence of diabetes compared to the rest of the nation (4). One of the most common approaches to explore the association between diabetes and genetic traits is the genetic polymorphism study on the susceptibility to T2DM (5).
Sirtuins comprise a group of protein deacetylases that rely on nicotinamide adenine dinucleotide (NAD) for their enzymatic activity (6), and these proteins are encoded by sirtuin genes (SIRT1 to SIRT7), with the SIRT1 (silent information regulator 1) gene being the first identified and most studied member of the family (1,7). Accumulating evidence shows that SIRT1 may produce antidiabetic effects by regulating insulin release, sensitivity, and controlling inflammatory responses, and it could serve as a potential target for the treatment of T2DM (6,9).
The single-nucleotide polymorphism (SNP) rs3758391 is located in the promoter region of the SIRT1 gene, and recent studies have explored its impact on the risk of developing T2DM; however, the findings vary with different populations (3,10). Therefore, the goal of this study is to determine the genomic association of SNP rs3758391 in the SIRT1 gene with T2DM in the population of the Appalachian area.
Materials and Methods
This case-control research included 38 participants diagnosed with type 2 diabetes mellitus (T2DM) and 43 non-diabetic individuals as the control group. The subjects were enrolled from rural health clinics in central Appalachia between January 2025 and May 2025. The T2DM diagnosis was based on the International Diabetes Federation (IDF) guidelines, which define T2DM as a fasting blood glucose level of at least 6.1 mmol/L or an HbA1c value of 6.5% or higher. Exclusion criteria included prediabetic conditions, pregnancy, T1DM, chronic liver and kidney diseases, cancers, and autoimmune disorders. Ethical approval for the research was granted by the Appalachian College of Pharmacy’s ethical review board (IRB# 2024-001). Participants signed an informed consent form before their inclusion in the study. Anthropometric measurements were gathered from personal interviews.
Genotyping Using the Restriction Fragment Length Polymorphism (RFLP) Technique
The rs3758391 variant was genotyped using the polymerase chain reaction–restriction fragment length polymorphism (PCR-RFLP) method. As described in a previous study, the primers used were as follows: Forward, 5′‐GTCACGCAGGTAATTGATGCAG‐3′, and Reverse, 5′‐GGCTTAGTGGAAAGCCCTTC‐3′ (11). The PCR amplification was carried out in a total reaction volume of 20 µL, which consisted of 100 ng of the DNA, 1 µL each of forward and reverse primers, 10 µL of 2x PCR Master Mix, and the final volume made up using nuclease-free water. The DNA amplification protocol consisted of denaturing at 94°C for 3 minutes initially, followed by 30 cycles of 94°C for 30 seconds denaturation, 57°C for 30 seconds primer annealing, and 72°C for 30 seconds for extension. The final extension follows at 72°C for 5 minutes. A 1.5% agarose gel stained with ethidium bromide was used to examine the amplified PCR products that were separated to ensure a length of 241 base pairs. Once the size of the PCR product was verified, 10 µL was digested at 37°C for one hour using the restriction enzyme NlaIII (New England Biolabs, USA). A 2% ethidium bromide-stained agarose gel was used to visualize the digested fragments, and a 100-base pair DNA ladder was used to estimate the sizes. The TT genotype displayed 148 bp and 93 bp bands, the CC genotype showed a 241 bp fragment, and the CT genotype produced 241 bp, 148 bp, and 93 bp.
Results and Discussion
The current study described that the demographic and anthropometric characteristics of T2DM patients and healthy controls were statistically different, which included age, gender, body mass index (BMI), fasting blood glucose (FBG), glycated hemoglobin or HbA1c, high-density lipoprotein (HDL), systolic blood pressure (SBP), and triglycerides (TG) (Table 1). The frequency distribution of the SIRT1 rs3758391 C/T genotypes and alleles was compared to 38 T2DM patients and 43 controls.
Table 1: Anthropometric and clinical parameters of T2DM patients and healthy controls.
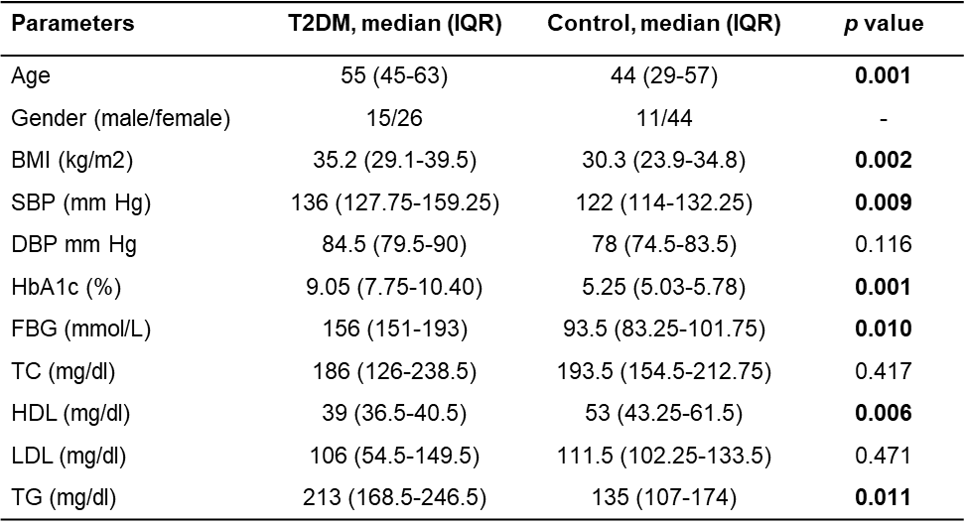
Note: The Mann‐Whitney U test was used to compare the continuous variables between the T2DM and control groups. p < 0.05 was considered statistically significant and was made bold. Abbreviations: Interquartile range (IQR), diastolic blood pressure (DBP), systolic blood pressure (SBP), low-density lipoprotein (LDL), total cholesterol (TC), high-density lipoprotein (HDL), total cholesterol (TC), Triglyceride (TG), fasting blood glucose (FBG).
The alteration in the SIRT1 gene has been associated with an increased likelihood of T2DM, underscoring its involvement in disease susceptibility (12). Studies suggest that activation of SIRT1 enhances insulin release and boosts glucose regulation by influencing some important metabolic routes, such as AMPK and PGC-1α signaling pathways (13). In animal models, compounds that activate SIRT1, such as resveratrol, have demonstrated therapeutic potential by improving blood glucose regulation and reducing inflammation (14), while human studies support findings linking the polymorphism of the SIRT1 gene to increased risk of T2DM (15).
The association of SIRT1 polymorphism with T2DM was represented by the odds ratio (OR) with 95% confidence interval (95% CI) (Table 2). Data from our small-scale genome-wide association study (GWAS) between rs3758391 polymorphism and increased risk of T2DM did not show any significant relationship under co-dominant TT vs CC (OR=2.625, 95% CI=0.46-15.11, P=0.27) & CT vs CC (OR=1.26, 95% CI=0.50-3.15, P=0.62), recessive TT vs CC+CT (OR= 2.368, 95% CI= 0.43-12.99, P= 0.31) and allelic T vs C (OR=1.44, 95% CI=0.72-2.88, P= 0.29) genetic models.
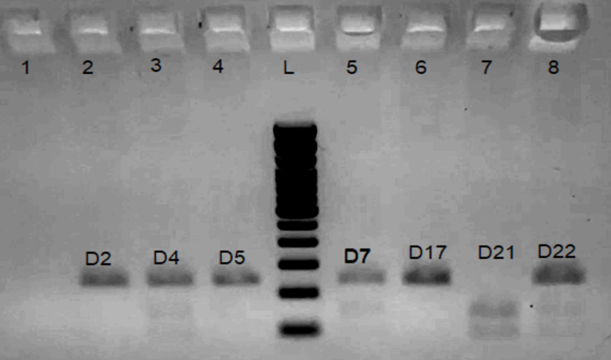
Figure 1: Restriction fragment length polymorphism analysis of rs3758391 C/T single-nucleotide polymorphism (SNP) using NlaIII enzyme. 100-bp ladder: Lane L; CC (241 bp): Lane 2, 4, 6; TT (148 bp and 93 bp): Lane 7; CT (241 bp, 148 bp, and 93 bp): Lane 3, 5, 8
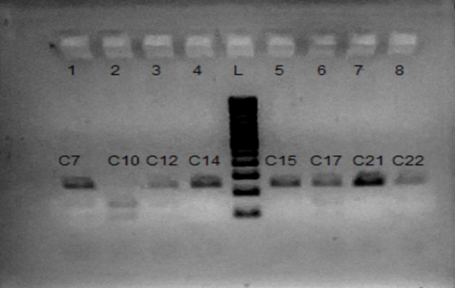
Figure. 2. Restriction fragment length polymorphism analysis of the rs3758391 C/T single-nucleotide polymorphism (SNP) using the NlaIII enzyme. 100-bp ladder: Lane L; CC (241 bp): Lane 4, 5, 6; TT (148 bp and 93 bp): Lane 2; CT (241 bp, 148 bp, and 93 bp): Lane 1, 3, 6, 8
Table 2: Genotype and allelic frequency of SIRT1 rs3758391 polymorphism in T2DM and healthy controls

Conclusion
The current research study showed no significant association between rs3758391 and T2DM in the Appalachian population. However, the number of cases and controls is a limitation of this study. Our recommendation is to make a set of replicates with larger sample sizes and in other ethnic groups that might generate more conclusive data. Moreover, the genomic association of the deadliest diseases has almost no cure to date. Therefore, the treatment of T2DM must consist of teaching patients regarding dietary knowledge, as well as regular monitoring and treatment with conventional and natural medicine will be the best approaches to minimize the burden of high mortality rates.
Author Contributions
Md. Mazharul Islam Chowdhury, Ph.D: resources, conceptualization, investigation, formal analysis, funding acquisition, methodology, visualization, writing–original draft, supervision. Tauna Gulley, Ph.D: collection of samples, data curation. Francis Obinna Asogwa: resources, writing–review, data curation, and editing. Randy Mullins, PharmD: project administration.
Acknowledgments
The authors are grateful to Susan Mayhew, PharmD, Dean, Appalachian College of Pharmacy, and Mr. McGlothlin for their support in our project to be done. This work was financially supported by a research grant, IRB# 2024-001
Conflicts of Interest
The authors declare no conflicts of interest.
Data Availability Statement
All authors have read and approved the final version of the manuscript. The corresponding author had full access to all the data in this study and takes complete responsibility for the integrity of the data and the accuracy of the data analysis. The authors confirm that the data supporting the findings of this study are available within the article. The data that support the findings of this study are available from the corresponding author upon reasonable request.
Transparency Statement
The lead author, Md. Mazharul Islam Chowdhury, Ph.D, affirms that this manuscript is an honest, accurate, and transparent account of the study being reported; that no important aspects of the study have been omitted; and that any discrepancies from the study as planned (and, if relevant, registered) have been explained.
References
- Naseri R, Khalili F, Rahimi Z, Yari K, Rezaei M. Protective role of SIRT1 (rs3758391 T > C) polymorphism against T2DM and its complications: Influence on GPx activity. Health Sci Rep. 2024;7(11):e70106.
- Ong KL, Stafford LK, McLaughlin SA, Boyko EJ, Vollset SE, Smith AE, et al. Global, regional, and national burden of diabetes from 1990 to 2021, with projections of prevalence to 2050: a systematic analysis for the Global Burden of Disease Study 2021. The Lancet. 2023 Jul 15;402(10397):203–34.
- Ahmed R, Safa MR, Zahid ZI, Chowdhury MdMI, Hasan ABMK, Mostaid MdS, et al. Association of SIRT1 rs3758391 Polymorphism With T2DM in Bangladeshi Population: Evidence From a Case-Control Study and Meta-Analysis. Health Sci Rep. 2025;8(2):e70495.
- Barker L, Gerzoff R, Crespo R, Shrewsberry M. Age at diagnosis of diabetes in Appalachia. Popul Health Metr. 2011 Sep 30;9(1):54.
- Kreienkamp RJ, Voight BF, Gloyn AL, Udler MS. Genetics of Type 2 Diabetes. In: Lawrence JM, Casagrande SS, Herman WH, Wexler DJ, Cefalu WT, editors. Diabetes in America [Internet]. Bethesda (MD): National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK); 2023 [cited 2025 Jul 8]. Available from: http://www.ncbi.nlm.nih.gov/books/NBK597726/
- Donmez G, Guarente L. Aging and disease: connections to sirtuins. Aging Cell. 2010;9(2):285–90.
- Dardano A, Lucchesi D, Garofolo M, Gualdani E, Falcetta P, Sancho Bornez V, et al. SIRT1 rs7896005 polymorphism affects major vascular outcomes, not all-cause mortality, in Caucasians with type 2 diabetes: A 13-year observational study. Diabetes Metab Res Rev. 2022;38(4):e3523.
- Cao Y, Jiang X, Ma H, Wang Y, Xue P, Liu Y. SIRT1 and insulin resistance. J Diabetes Complications. 2016 Jan 1;30(1):178–83.
- Kitada M, Ogura Y, Monno I, Koya D. Sirtuins and Type 2 Diabetes: Role in Inflammation, Oxidative Stress, and Mitochondrial Function. Front Endocrinol [Internet]. 2019 Mar 27 [cited 2025 Jun 25];10. Available from: https://www.frontiersin.org/journals/endocrinology/articles/10.3389/fendo.2019.00187/full
- Ramírez Á, Hernández M, Suárez-Sánchez R, Ortega C, Peralta J, Gómez J, et al. Type 2 diabetes–associated polymorphisms correlate with SIRT1 and TGF-β1 gene expression. Ann Hum Genet. 2020;84(2):185–94.
- Sadeghi MB, Nakhaee A, Saravani R, Sadeghi MH, Sargazi S, Nia MH. SIRT1 functional polymorphisms (rs12778366, rs3758391) as genetic biomarkers of susceptibility to type 2 diabetes mellitus in Iranians: a case-control study and computational analysis. Int J Diabetes Dev Ctries. 2021 Jul 1;41(3):447–55.
- Kitada M, Koya D. SIRT1 in Type 2 Diabetes: Mechanisms and Therapeutic Potential. Diabetes Metab J. 2013 Oct;37(5):315–25.
- Zhang T, Kraus WL. SIRT1-dependent regulation of chromatin and transcription: linking NAD(+) metabolism and signaling to the control of cellular functions. Biochim Biophys Acta. 2010 Aug;1804(8):1666–75.
- Kitada M, Ogura Y, Monno I, Koya D. Sirtuins and Type 2 Diabetes: Role in Inflammation, Oxidative Stress, and Mitochondrial Function. Front Endocrinol. 2019 Mar 27;10:187.
- ZHUANPING Z, RIFANG L, QING C, SIDONG C. The Association between SIRT1 Genetic Variation and Type 2 Diabetes Mellitus Is Influenced by Dietary Intake in Elderly Chinese. Iran J Public Health. 2018 Sep;47(9):1272–80.
Note
Copyright © 2026 Chowdhury et al. Place of Publication: PSciP Publishing LLC, Oakwood, VA, USA.
